Science | Interactions between nascent proteins translated by adjacent ribosomes drive homomer assembly

Most proteins in the cell need to be combined with other proteins and form protein complexes to function. However, this is not an easy task, because a protein needs to accurately find other subunits of the complex in the cell environment, and avoid non-specific binding with other proteins.
Current research shows that in order to ensure the efficient and correct assembly of protein complexes, the strategies adopted by cells include: precisely controlling the expression of each protein to match the stoichiometry between the subunits of the complex; using molecular chaperones to guide protein binding; Co-localization of the synthesis of different subunit proteins; timely assembly of the translation products, etc. In these models, the folded protein is required to co-post assembly with its new partner subunit through free diffusion.
Prof.Günter Kramer and Prof.Bernd Bukau from the University of Heidelberg, Germany published a study in Science Interactions between nascent proteins translated by adjacent ribosomes drive homomer assembly. The study found that co-co assembly is a universal and efficient strategy for the assembly of nascent protein subunits into protein dimers.
In this study, the researchers proposed a very creative model: “Can a protein translated on the same mRNA be combined while being translated, and finally form an active protein dimer form?” This model can circumvent the problem of how effectively the protein that has been translated and folded can diffuse freely to the vicinity of the protein that needs to be bound.
To test this hypothesis, the researchers used nuclease to treat the cell lysate, and then separated the ribosomes by density gradient centrifugation. Under the treatment of nuclease, the ribosomes distributed on the same mRNA will separate due to the digestion of the mRNA; and if the new protein translated by the two ribosomes is combined, these two ribosomes form dimeric ribosomes after nuclease treatment.
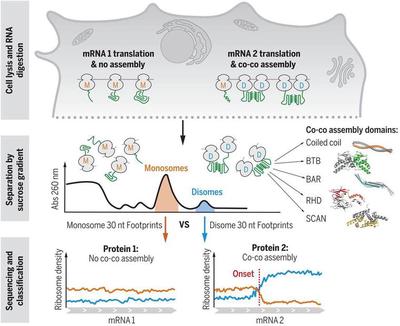
Indeed, through density gradient centrifugation, the researchers observed that some of the ribosomes formed dimers. So are these ribosomes connected by nascent proteins? Under the treatment of puromycin and a certain concentration of proteinase K, the researchers found that these dimeric ribosomes had restored the state of single ribosomes, indicating that they were indeed connected by nascent proteins.
So, what kind of protein is more inclined to adopt the mechanism of translation and assembly to form homodimers? The researchers identified a total of 829 high-confidence proteins using this strategy. Further analysis showed that these proteins contained specific domains, including Coiled coil, BTB, BAR, RHD, and SCAN, among which the N-terminal Coiled coil structure of the protein was the most universal.
In general, the study found a new way of protein assembly through ribosome separation and sequencing, called co-co assembly (Figure 1). This method is more efficient and accurate than the method of forming protein complexes by free diffusion, because the new peptide chains to be combined are closer to the adjacent ribosomes. At the same time, this way of protein assembly makes the two proteins forming the dimer come from the same mRNA, avoiding non-specific binding. The proposal of this model is of great significance for understanding the efficiency and correctness of protein assembly in organisms.
Reference
- Bertolini, M., Fenzl, K., Kats, I., Wruck, F., Tippmann, F., Schmitt, J., Auburger, J. J., Tans, S., Bukau, B., & Kramer, G. (2021). Interactions between nascent proteins translated by adjacent ribosomes drive homomer assembly. Science (New York, N.Y.), 371(6524), 57–64.
- BioArt Posts
- Li, G. W., Burkhardt, D., Gross, C., & Weissman, J. S. (2014). Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell, 157(3), 624–635.
- Shieh, Y. W., Minguez, P., Bork, P., Auburger, J. J., Guilbride, D. L., Kramer, G., & Bukau, B. (2015). Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science (New York, N.Y.), 350(6261), 678–680.
- Hampoelz, B., Schwarz, A., Ronchi, P., Bragulat-Teixidor, H., Tischer, C., Gaspar, I., Ephrussi, A., Schwab, Y., & Beck, M. (2019). Nuclear Pores Assemble from Nucleoporin Condensates During Oogenesis. Cell, 179(3), 671–686.e17.
- Shiber, A., Döring, K., Friedrich, U., Klann, K., Merker, D., Zedan, M., Tippmann, F., Kramer, G., & Bukau, B. (2018). Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature, 561(7722), 268–272.