Nature Communications | Network medicine for disease module identification and drug repurposing with the NeDRex platform

Standing on the shoulders of giants
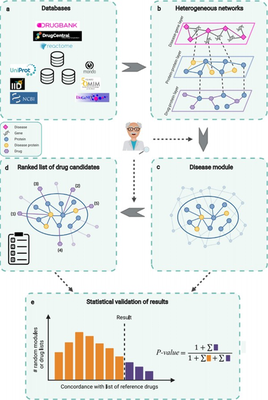
Studies have shown that genes associated with disease are not randomly distributed throughout biological networks. Instead, they are often located in disease modules, small subnetworks of association mechanisms that can be linked to the phenotype. One of the guiding paradigms for network-based drug repurposing is that disease can be considered as a perturbation of these modules. Thus, potentially repurposable drugs can be identified in the computer in three steps as follows:
-
Integrating data from multiple biomedical databases related to the given task to construct heterogeneous biological networks.
-
Mining the constructed biological network to obtain the disease modules associated with this disease.
-
Extraction of a prioritization table of drugs in disease modules where the drug’s known targets are contained in or located near the extracted disease module.
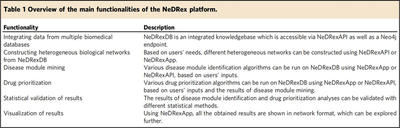
This paper presents an interactive, user-friendly platform that helps scientists and clinicians with non-computer backgrounds to mine different layers of a large heterogeneous biological network, the NeDRexDB knowledgebase. NeDRex provides users with multiple web-based methods to obtain disease modules associated with the disease under study and to directly or indirectly sequence drugs against the disease modules. NeDRex can also prioritize approved drugs to skip the preclinical study phase and go directly to clinical trials, speeding up the drug development process. Thanks to the expert engagement model, researchers from the biomedical sciences can leverage their domain knowledge in different nodes of the workflow, for example, by using the platform to filter disease genes already provided, or by using their own gene sets as the start of the algorithm.
NeDRex is built of three main components: a knowledgebase (NeDRexDB, available at http://neo4j.nedrex.net/ and https://api.nedrex.net/ ), a Cytoscape app (NeDRexApp, available at https://apps.cytoscape.org/apps/nedrex ), and an API (NeDRex- API, available at https://api.nedrex.net/ ).
Flowchart
General review of the NeDRex platform:
Functions

Reference
- Sadegh S, Skelton J, Anastasi E, et al. Network medicine for disease module identification and drug repurposing with the NeDRex platform. Nat Commun. 2021;12(1):6848.