Science | Further refinement of the human genome
Filling the last 8% of the human genome

Twenty years ago, Science and Nature first published the human genome sequences completed by Celera Genetics and the Human Genome Project (HGP), respectively, which were incomplete and full of errors. The Human Genome Project has since completed most of the sequence assembly and has updated it several times since then, and the version of the human genome is now GRCh38.p14 (GRCh38).
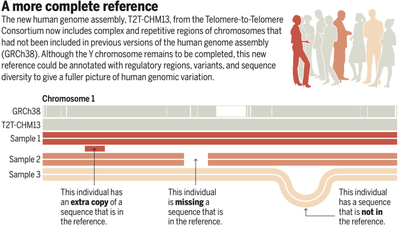
GRCh38 is missing about 8% of the human genome, with millions of bases unknown, indicated by the letter “N”; 169 significant repetitive sequences were not successfully spliced; and a significant portion of the sequence was difficult to analyze and assemble. The short arms of the proximal chromosomal mitoses, the mitoses and several repetitive euchromatin regions with important biological functions have also not been resolved and can only be represented by pattern sequences. This information is relevant to numerous human diseases, and the deciphered assembly of these important sequences has not been achieved due to the limitation of sequencing technology.
The completed human genome T2T-CHM13 is haploid, and the sequenced samples were derived from staph-derived cell lines, ensuring that all sequencing was from identical haploids. Many biases in the GRCh38 version were corrected, and a large number of unknown significant sequences were resolved.
The current release of the genome sequence still has a shortcoming: T2T-CHM13 does not have a Y chromosome sequence due to sequencing samples from a haploid-derived cell line, which scientists will address subsequently. Based on the previous version of GRCh38 there are many important genomic annotations that need to be integrated into T2T-CHM13, which is the key to exploit the genomic sequence. The Genome Consortium will conduct more analytical interpretations of the human genome, and this information will better help us understand ourselves and serve scientific research.