WIREs computational molecular science | Along the allostery stream - Recent advances in computational methods for allosteric drug discovery

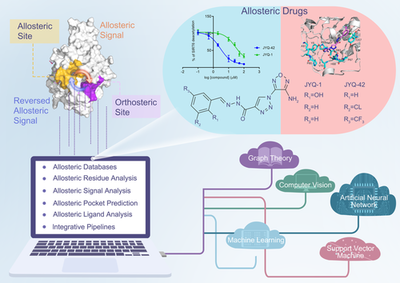
Allostery is a common phenomenon in living proteins: small perturbations (e.g., ligand binding, residue mutations, or post-translational modifications) trigger an allosteric site far from the orthosteric site in the topology to fine-tune the orthosteric site. Allosteric phenomena are associated with many physiological and pathological conditions and can precisely up- and down-regulate protein function through different allosteric sites, but all previously discovered allosteric drugs have been discovered by chance through large-scale experimental screening. With the establishment and continuous development of the ASD (http://mdl.shsmu.edu.cn/ASD), the design of drugs from allosteric conformations has become a new idea in drug discovery. Meanwhile, significant advances in biophysics, especially in its branch of structural biology, have increased the understanding of the spatial characterization of allosteric effects. These multidimensional data provide an opportunity for the development of computational science methods for allosteric drug discovery. This paper illustrates how to establish a multidimensional “atom-residue-site-molecule-network” computational method for allosteric drug discovery from big data, and use it effectively for allosteric drug discovery.
Rich and high-quality data are the basis for algorithm training and model optimization, as well as for further evaluation and validation, and are an indispensable part of computational method development. This paper describes the design of the data framework ASD, proposes the composition dimensions and criteria of allosteric data for parsing the full range of information related to allosteric events, and creates ASBench (http://mdl.shsmu.edu.cn/asbench) for allosteric algorithm development feature data, based on which the characterization of allosteric residues, the pathological significance of allosteric sites and the bidirectional transmission effect of allosteric signals within proteins are analyzed to develop an in-depth understanding of the allosteric molecular mechanism. They have developed a deeper understanding of the molecular mechanism of allostery. Further, they compared the methods of allosteric sites and allosteric regulatory molecules, and suggested that most of the allosteric sites of regulatory proteins are not intuitively present on the surface of crystal structures, but are mainly hidden in temporary sites in different conformational states of proteins, which inspired them to develop methods for the identification of “cryptic allosteric sites” and their application in the drug discovery of difficult-to-target proteins.
As a result, the authors propose that computational approaches to drug development using allosteric therapeutics will become one of the standard practices in drug development methods in the modern medical era. Complemented by approaches from other areas of biology, big data-driven computational methods for allosteric therapeutics can become a powerful weapon in our efforts to overcome various difficult targets and open up a new pathway for First-in-class drug development advances.
Reference
- Ni D, Chai Z, Wang Y, Li M, Yu Z, Liu Y, et al. Along the allostery stream: Recent advances in computational methods for allosteric drug discovery. WIREs Comput Mol Sci. 2021;e1585.
- Molecular Design Laboratory.